The iBioGen Toolkit is conceived as a collection of resources for biodiversity genomics:
- Standardized field and lab protocols for next generation biodiversity monitoring
- Open-source pipelines for processing high throughput sequence datasets
- Software for analysis of community-level genetic data
- Online resources for training
- Review and opinion papers that will promote methodological unification and theoretical synthesis in the field of biodiversity genomics
Due to the modular nature of the toolkit, this section will be constantly updated, following the completion of each tool. Below we provide information and links to already published online resources and articles, ready-to-use software and relevant protocols.
- Standardized field and lab protocols for next generation biodiversity monitoring
- Open-source pipelines for processing high throughput sequence datasets
- Software for analysis of community-level genetic data
- Online resources for training
- Review and opinion papers that will promote methodological unification and theoretical synthesis in the field of biodiversity genomics
Due to the modular nature of the toolkit, this section will be constantly updated, following the completion of each tool. Below we provide information and links to already published online resources and articles, ready-to-use software and relevant protocols.
Bioinformatic Methods for Biodiversity Metabarcoding
|
A free online self-training resource on bioinformatics methods for biodiversity metabarcoding. It provides a step-by-step guide on how to go from raw sequence reads to the generation of biodiversity data for a wide range of applications. It was produced in the context of iBioGen's online workshop Theory and practice in metabarcoding for biodiversity.
|
metaMATE
|
This software is designed to form part of a metabarcoding pipeline that uses a mitochondrial coding gene, such as Cox1, to filter out nuclear mitochondrial pseudogenes. It does this by identifying control sequences then evaluating the effects of abundance thresholds over different dataset partitioning strategies on the rates of valid read recovery and invalid read removal. The software allows careful decisions on the adequate metabarcoding filtering strategy for specific research objectives, improving the reliability of intraspecific genetic information derived from metabarcoding data.
|
'iBioGen' R package
|
Software for the study of island biodiversity dynamics from genomic data. It is composed of a suite of tools for studying ecological, evolutionary, and multi-scale processes within island communities.
|
PROTOCOL HARMONISATION OF METABARCODE DATA GENERATION
|
Harmonised implementation of metabarcoding protocols, is both timely and essential. iBioGen proposes a modular framework for data generation, which could offer a pathway to navigate the complex structure of terrestrial metazoan biodiversity.
|
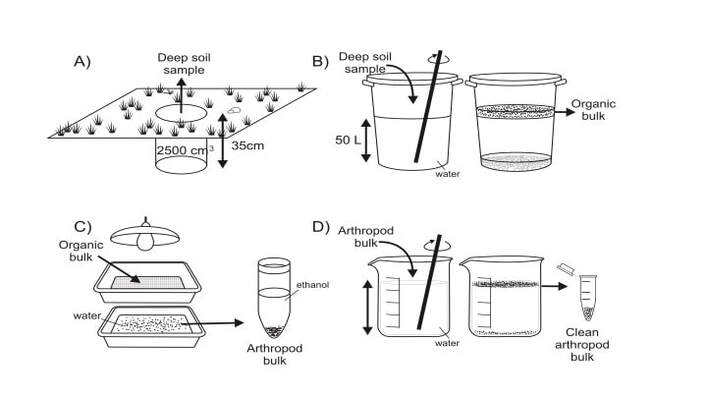
SOIL EXTRACTION
|
For specimen extractions from soil samples we follow the Flotation – Berlese – Flotation protocol of Arribas et al. (Methods in Ecology and Evolution, 2016, 7, 1071-1081).
Specimens are extracted from the soil matrix by flotation in a large water container, after which floating debris is subjected to drying in Berlese trays. Subsequently, the collected specimens are freed from remaining debris in a second round of flotation. For more details, please download the Arribas et al. (2016) article below. |