All public deliverables of the iBioGen project will be published here, when they become available.
Deliverable 1.4 - Events Year 3
Deliverable 1.4 presents the main activities and events organised during the period September 2020 – February 2022, which covers the third year of the project plus the 6 month extension. The activities include three annual meetings, an online training of University of Cyprus (UCY) students, two staff exchange visits, the organisation of an international online workshop on bioinformatic analyses of metabarcoding data, the organization of two online working groups on protocol unification and on island Genomic Observatories, the organization of two seminars to stakeholders and the participation in Researchers night 2020. Additionally, three videos were created, an educational brochure, a permanent exhibit and four educational programs for the Environmental Education Centres in Cyprus. Statistics on the project’s online presence in social media are summarised and the publications generated during the third year of the project are listed.
For the detailed document of Deliverable 1.4 click here.
Deliverable 1.3 - Events Year 2
|
Deliverable 1.3 presents the main activities and events organised during the period September 2019 – August 2020, which covers the second year of the project. The activities include the annual review meeting of the project, a training visit of University of Cyprus (UCY) students to the Natural History Museum (NHM) in the UK, a staff exchange visit between UCY and NHM, the organisation of an International workshop on metagenetics in Cyprus, the organization of an international symposium on Genomic Observatories in Cyprus and the organisation of an online working group on Ecological-Evolutionary synthesis. In addition to these activities, statistics on the project’s online presence in social media are summarised and the publications and pieces of software generated during the second year of the project are listed.
|
For the detailed document of Deliverable 1.3 click here.
Deliverable 3.1 - Symposium opinion paper and online report
|
The Symposium “Next Generation Biodiversity Monitoring” took place on the 11th-13th of November 2019 at the University of Cyprus (UCY). Twenty-five international researchers, including evolutionary geneticists, ecologists, and biodiversity conservationists, participated in this symposium that was conceived to discuss how best to develop an integrative high throughput sequencing (HTS) framework for measuring and understanding global patterns of biodiversity. The universality, reliability, and efficiency of HTS data can potentially facilitate the seamless linking of data among species assemblages from different sites, at different hierarchical levels of diversity, for any taxonomic group and regardless of prior taxonomic knowledge. However, collective international efforts are required to optimally exploit the potential of site-based HTS data for global integration and synthesis, efforts that at present are limited to the microbial domain. This symposium was conceived to contribute to the development of an analogous strategy for the non-microbial terrestrial domain.
|
The symposium was organised in two main blocks. The first block on day one started with an overview of the iBioGen project and the main aims of the symposium were presented to all the participants. Subsequently, the invited participants gave presentations organised within four main sessions entitled: (i) Defining the Genomic Observatory (GO) concept in the era of HTS; (ii) GOs and current initiatives for biodiversity surveying and monitoring; (iii) Core data and measures from GOs and (iv) Linking GO data and synthesis of biodiversity patterns. This first day of the symposium was open to the public and a considerable number of local stakeholders and members from UCY attended. This initial day provided the invited participants the opportunity to share and discuss their research and experience on the use of genomic-based approaches for biodiversity measurement, thus generating a stimulating atmosphere on the topic to build on during the following two days of the symposium. The second and third days consisted in a second block of closed discussion sessions that were structured to discuss main challenges to be addressed: (i) a GO definition that builds optimally on the diversity of current initiatives for biodiversity surveying and monitoring; (ii) Core data and measures to be provided by GOs and the molecular tools to obtain them; (iii) Parameterising the definition of a GO network: an integrative and modular framework for data acquisition, and; (iv) designing a modular framework for data acquisition.
Discussion on the current state of site-based biodiversity science using DNA sequence information converged on the high potential of HTS approaches to shift from incidental observations toward site-based biodiversity recording. Discussion also centred on how the GO network concept proposed by Davies et al. (2012, 2014) could be adapted to catalyse such a shift. A consensus view emerged with regard to the need for a realistic appreciation of logistical and financial constraints in attempting to integrate the three principal axes of space, time, and genome, for the implementation of the conceptual GO framework of Davies et al. (2014, 2012). In this sense, the discussion within the symposium converged on the opinion that an effective, spatially-led terrestrial GO network should place importance on maximising the global distribution of sampling sites and that these should be strategically united under the umbrella of the single-locus, per taxon metabarcoding approach. Within the spatially-led terrestrial GO network that we propose, we also envision that both the temporal and genomic axes can be more deeply sampled at particular sites, consistent with the idea of ‘model ecosystems’, and we refer to them as ‘SuperGOs’. Based on this framework, we agreed that optimal comparison and integration among independently generated community datasets would be best served by the establishment of harmonised molecular approaches for site-based biodiversity surveys. The remainder of the symposium was thus focussed on identifying how to collectively build an integrative framework for data generation.
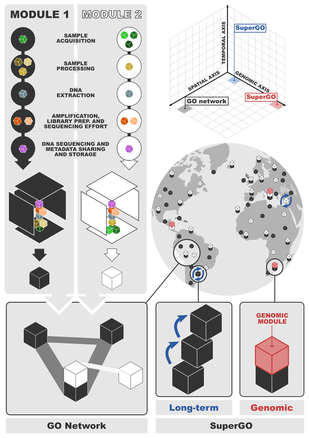
A spatially-led, non-centralised GO network would require bottom-up growth, and many existing international, national and regional projects or initiatives demonstrate the potential for this through broadly shared goals of generating site-based biodiversity data using metabarcoding. However, in addition to standardisation for reporting taxonomic, genomic and metadata, we also see the need for both procedures and best practices for the generation of site-based metabarcoding data to be similarly standardised. We discussed the potentially complex sampling requirements that would typically be needed to capture all non-microbial fractions of terrestrial diversity, and concluded that a “modular” structure for data acquisition could be the most promising approach. Modules, as basic building blocks of the framework, can provide simple, integrated and interoperable procedures for site-based characterisation of extensive biodiversity fractions. An appropriate module design would comprise a minimum set of protocols (or sub-modules) across the sequence of steps from sample acquisition to sequencing, which would facilitate the generation of inventory data across a wide taxonomic spectrum (if possible, complete biotas), in a simultaneous, integrated and cost-efficient way. Biodiversity fractions for which (i) there is a high potential for global integration from site-based data, and (ii) metabarcoding protocols, datasets or sampling programs already exist (or are in an advanced stage of development), emerged as obvious candidates for an initial focus on module development. In the discussion, soil biodiversity and terrestrial arthropods emerged as ideal candidate modules to be further developed during a subsequent working group within the iBioGen project, due to the appreciable body of HTS work that has already been carried out on these biodiversity fractions.
The stimulating discussion and fruitful interactions among the participants during the symposium have resulted in a meeting review publication in the journal Molecular Ecology (https://doi.org/10.1111/mec.15797).
Deliverable 1.2 - Events Year 1
|
Deliverable 1.2 presents the main activities and events organised during the period September 2018 - August 2019, which covers the first year of the project. The activities include the kick-off meeting of the project, a training visit of the University of Cyprus students to the Natural History Museum in the United Kingdom, a staff exchange visit of the University of Cyprus personnel to the Instituto de Productos Naturales y Agrobiología (IPNA, CSIC) in Spain, the organization of an International symposium "Synthesizing island biodiversity theory for community-wide genetic data" as part of the Island Biology 2019 conference in France and the presentation of the project's objectives at the same international conference. In addition to these activities, statistics on the project's online presence in social media and other channels are summarized.
For the detailed document of Deliverable 1.2 click here |